The Digital Expression Explorer (DEE) project is one of the flagship projects for the Ziemann Computational Biology lab. Since 2015, the project has strived to deliver processed RNA-seq data into the public domain. If you have ever had to process this type of gene expression data you will know that it requires a great deal of computational power and expertise at using command-line tools. This means that most researchers, students and hobbyists are excluded from undertaking this sort of analysis. What DEE aims to do is undertake the data crunching for the many thousands of datasets online at NCBI GEO and make the processed files publicly available. These processed files contain summarised gene expression values for each gene in each sample, so instead of being >1GB per sample, the dataset is more like ~1MB per sample.
DEE version 2.0 was described in a GigaScience publication (Ziemann et al 2019) where we describe many improvements in the data analysis pipeline as well as methods to access data via the webpage and programmatically via R.
As of July 2021, DEE2 contains 1.5M RNA-seq datasets from ~1M samples, making it the largest such dataset in the public domain. DEE2 is therefore an important piece of genomics infrastructure, improving reuse of public RNA-seq data, feeding innovation into gene expression networks and machine learning. We continue to add new datasets to DEE2 thanks to the support of Deakin University, NCI/MASSIVE and Nectar Research Cloud for compute resources.
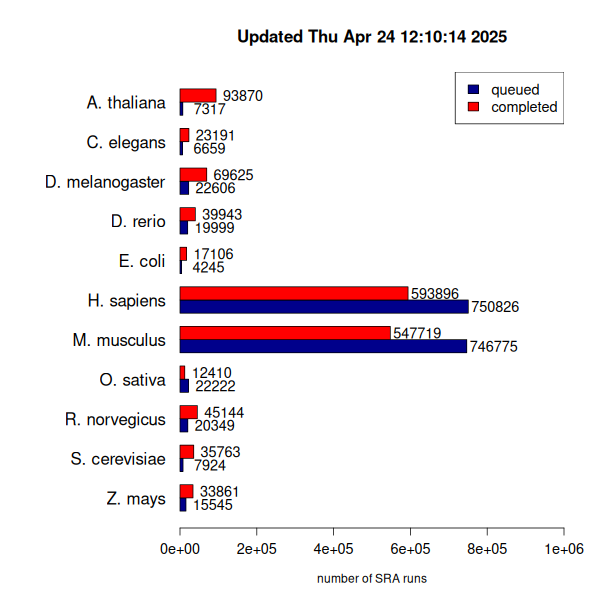
Visit http://dee2.io to access the dataset.
Further reading
Ziemann, M., Kaspi, A. and El-Osta, A. (2019) “Digital expression explorer 2: a repository of uniformly processed RNA sequencing data,” GigaScience, 8(4). doi: 10.1093/gigascience/giz022.
Ziemann M, Kaspi A (2021). getDEE2: Programmatic access to the DEE2 RNA expression dataset. R package version 1.2.0, https://github.com/markziemann/getDEE2.
Some of the projects that have used DEE2 data
Berry, A. S. F. et al. (2021) “An open-source toolkit to expand bioinformatics training in infectious diseases,” mBio, p. e0121421.
Mansouri, N. et al. (2019) “Mesenchymal stromal cell exosomes prevent and revert experimental pulmonary fibrosis through modulation of monocyte phenotypes,” JCI insight, 4(21). doi: 10.1172/jci.insight.128060.
Russkikh, N. et al. (2020) “Style transfer with variational autoencoders is a promising approach to RNA-Seq data harmonization and analysis,” Bioinformatics (Oxford, England), 36(20), pp. 5076–5085.
Yang, L. et al. (2020) “A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids,” Cell stem cell, 27(1), pp. 125-136.e7.
Zrimec, J. et al. (2020) “Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure,” Nature communications, 11(1), p. 6141.