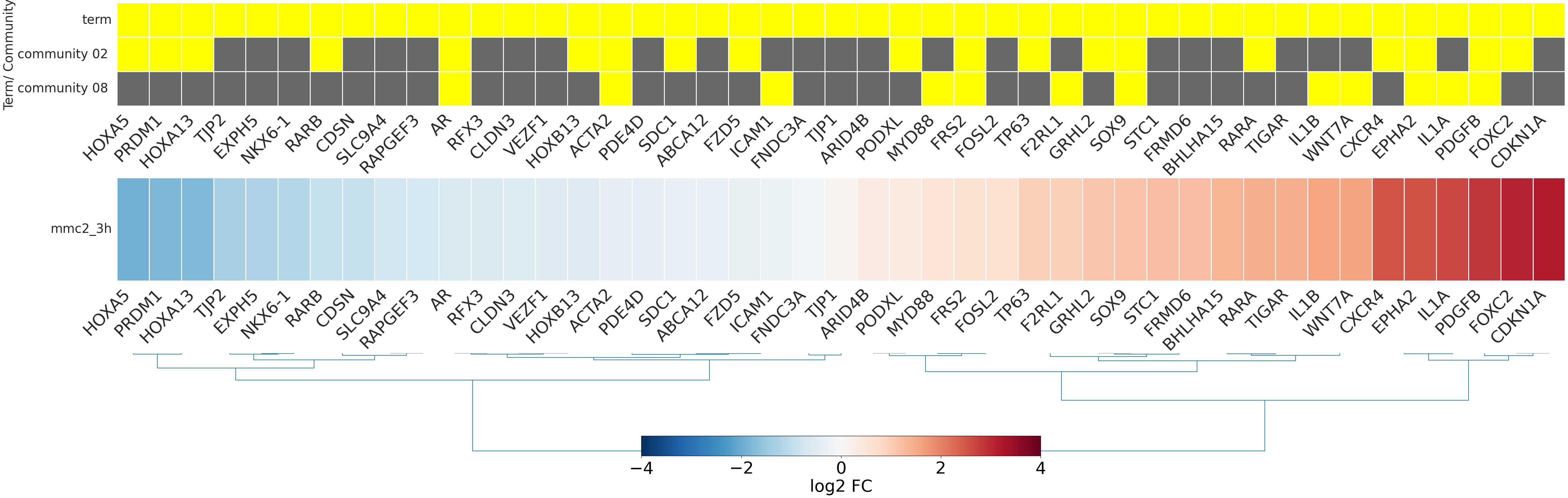
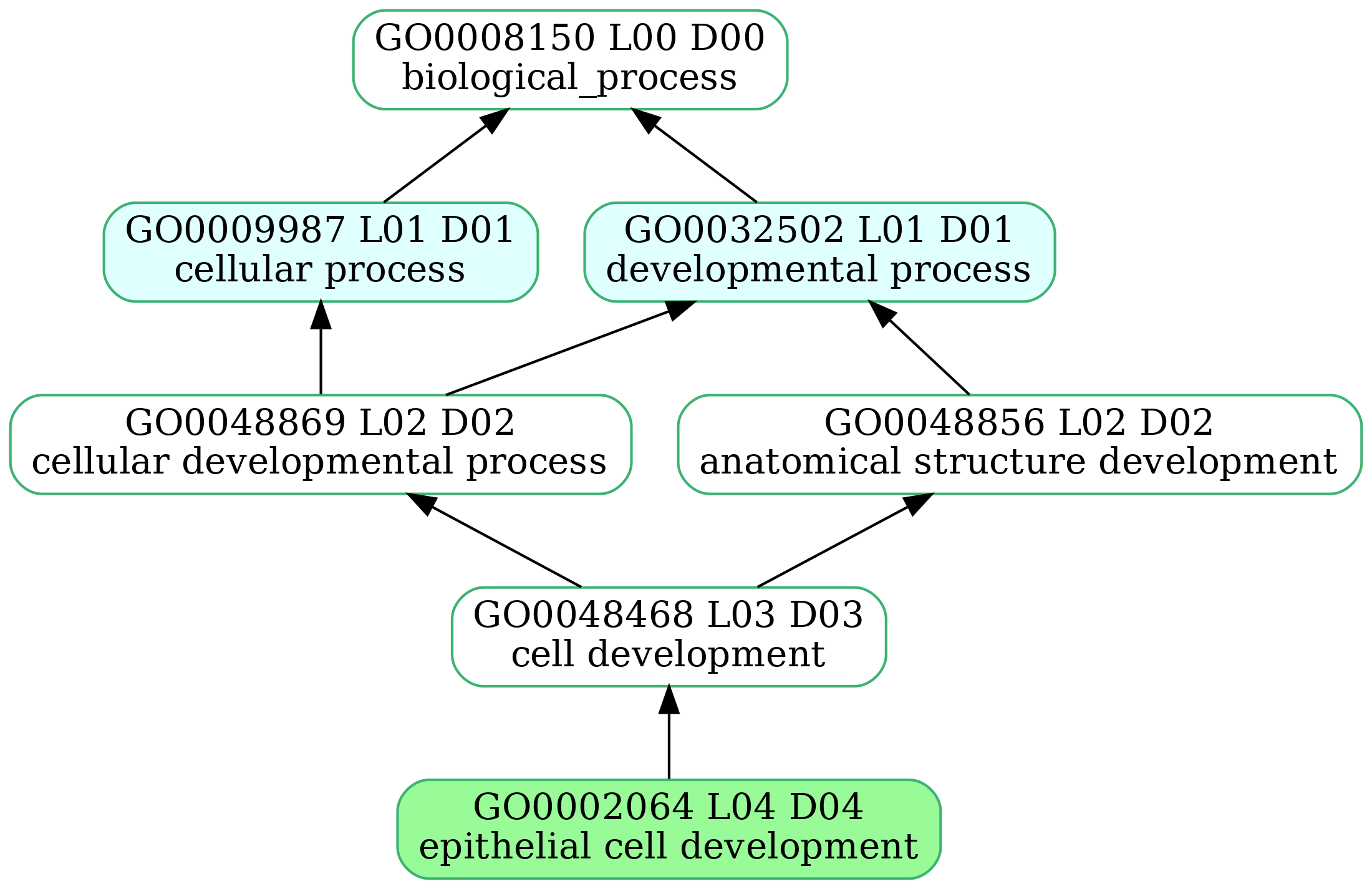
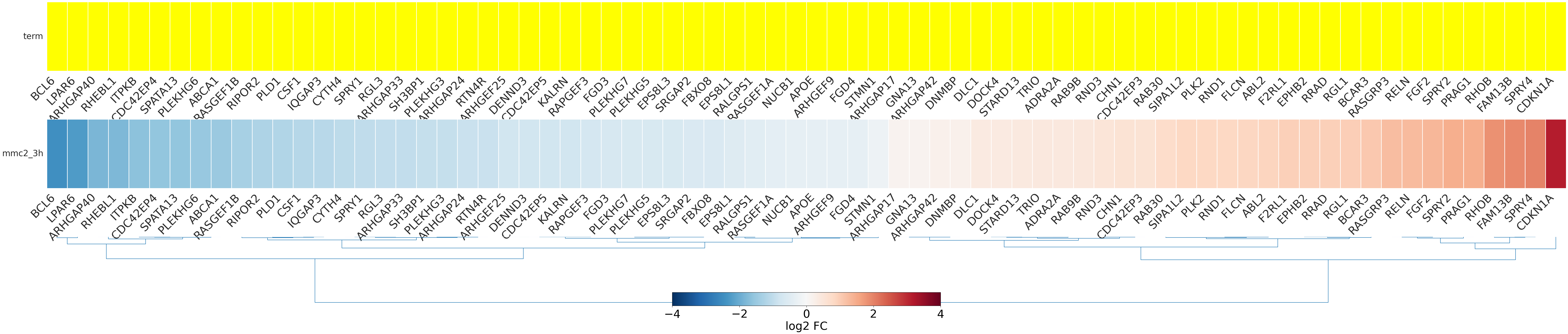
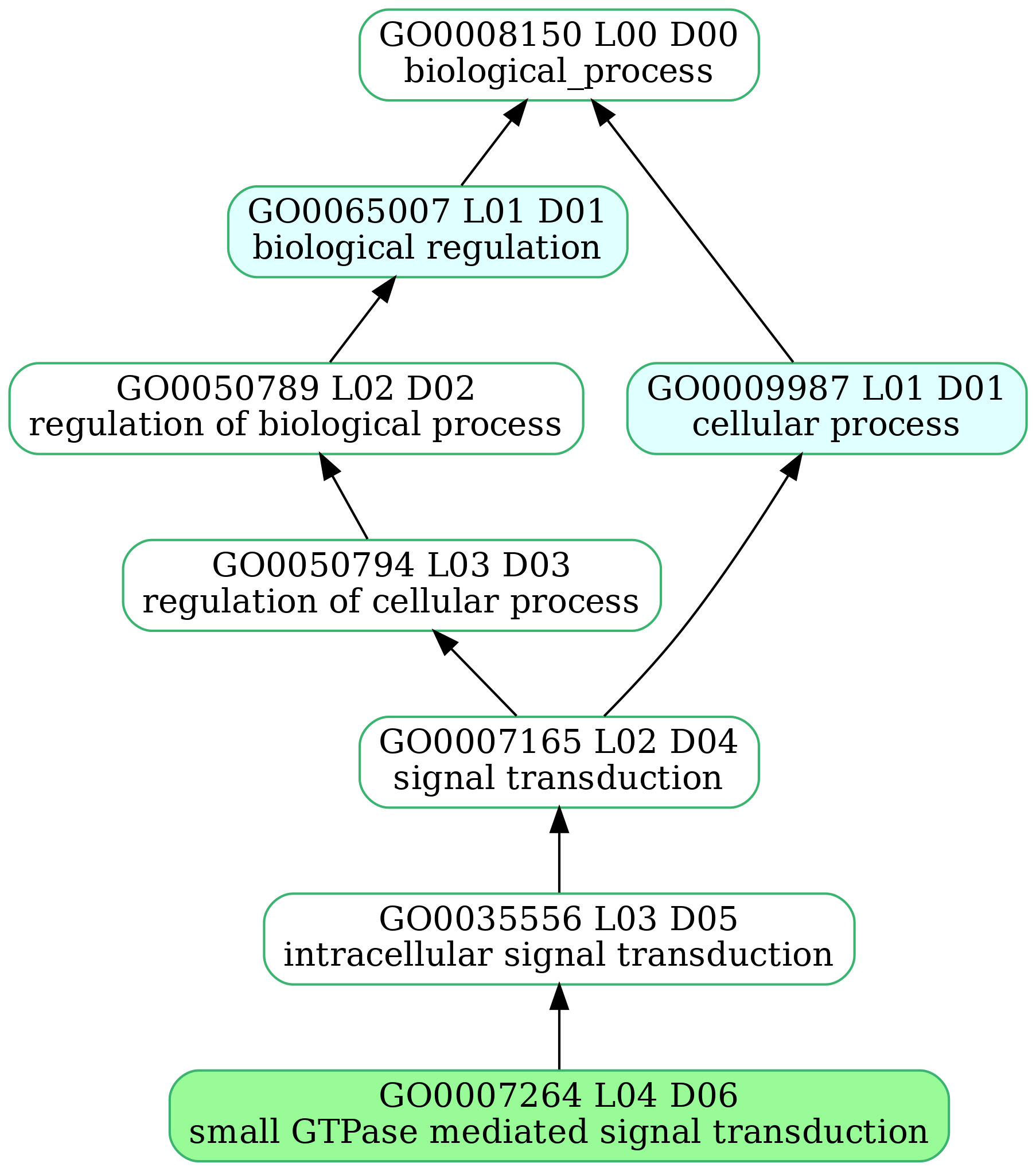
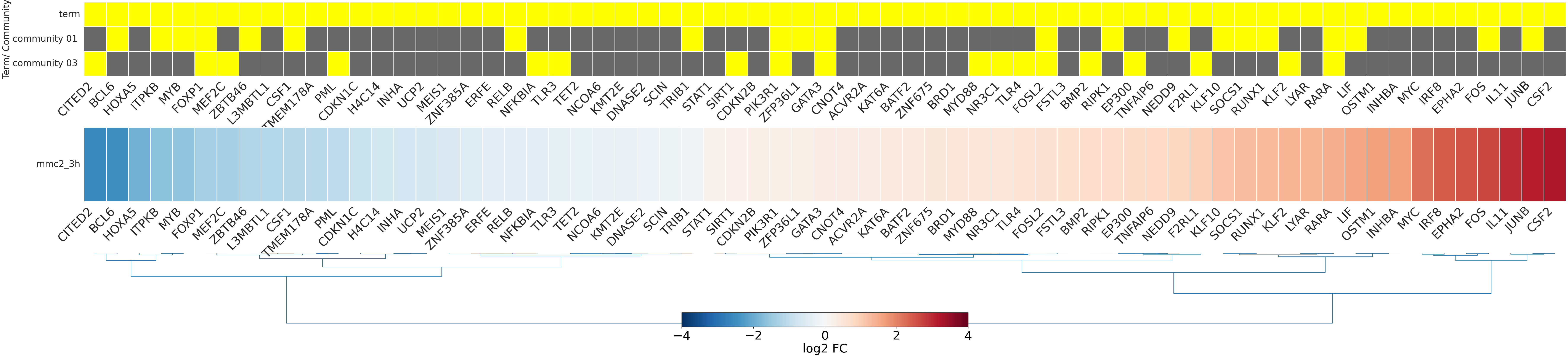
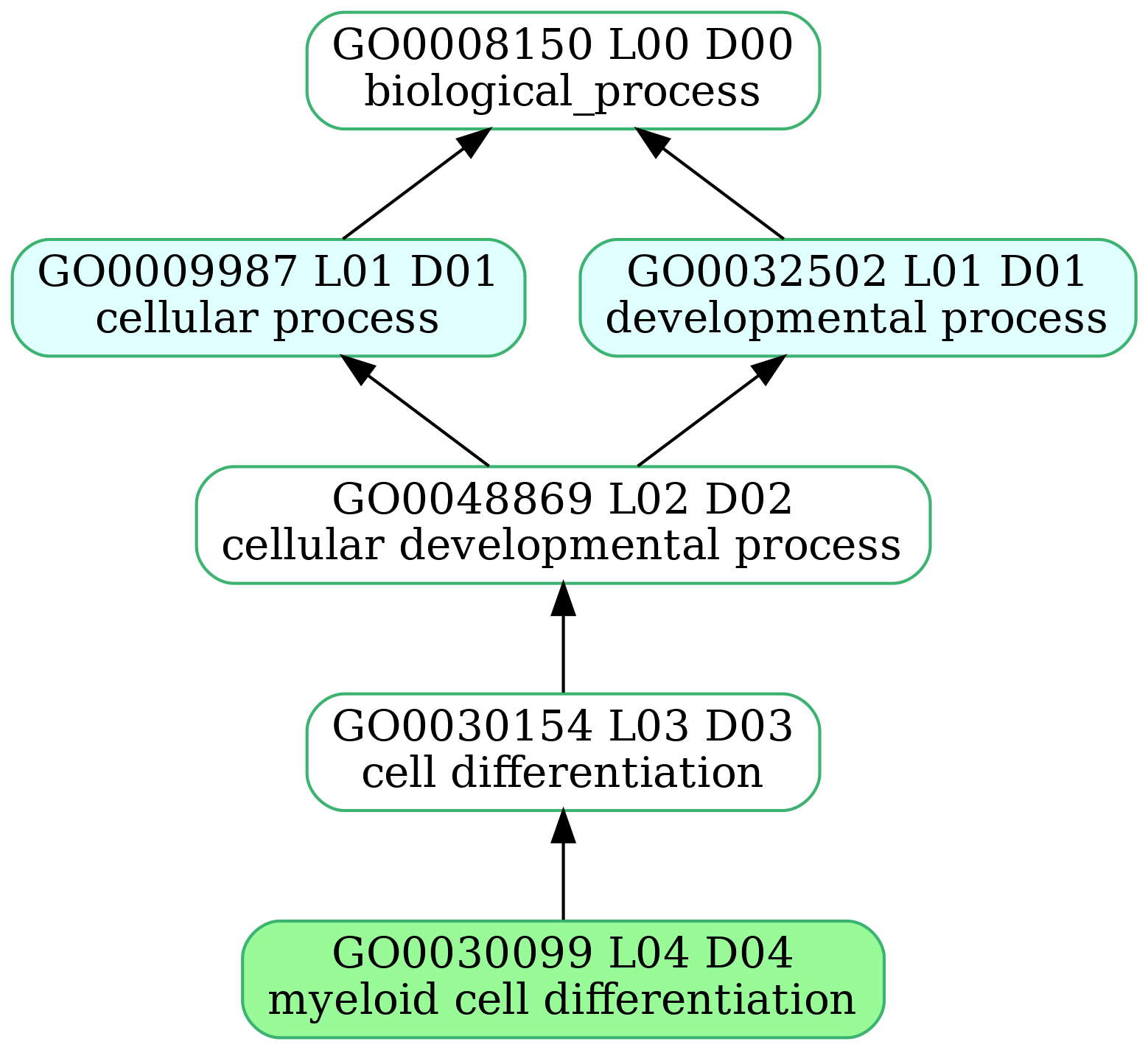
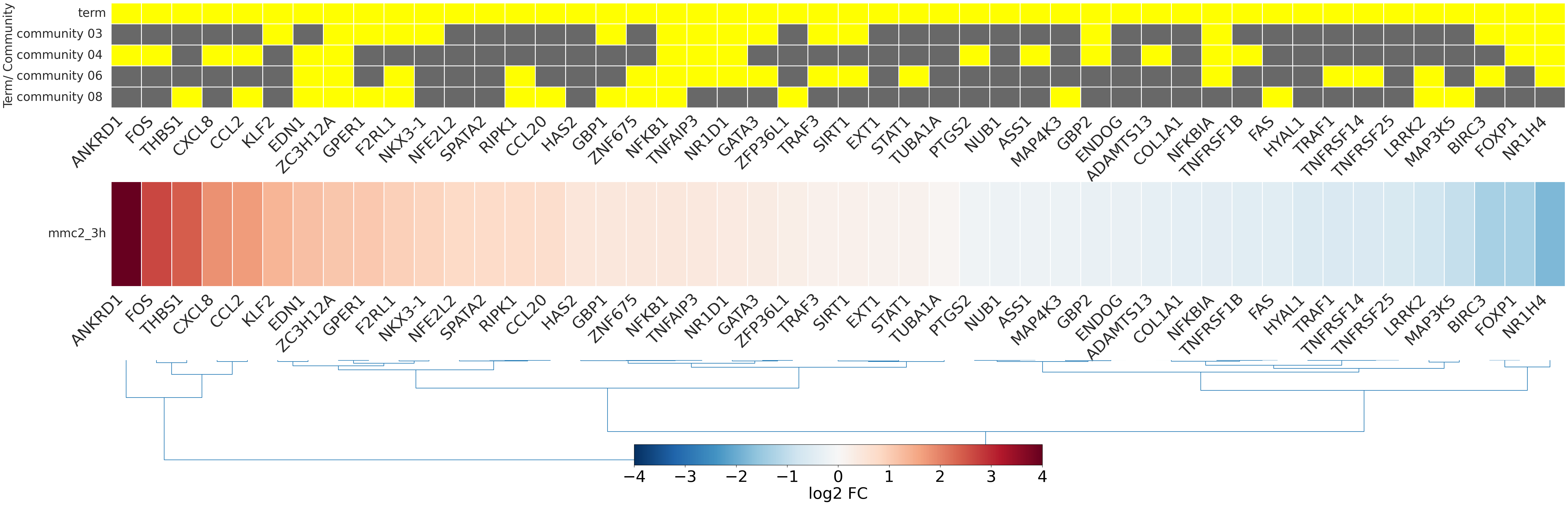
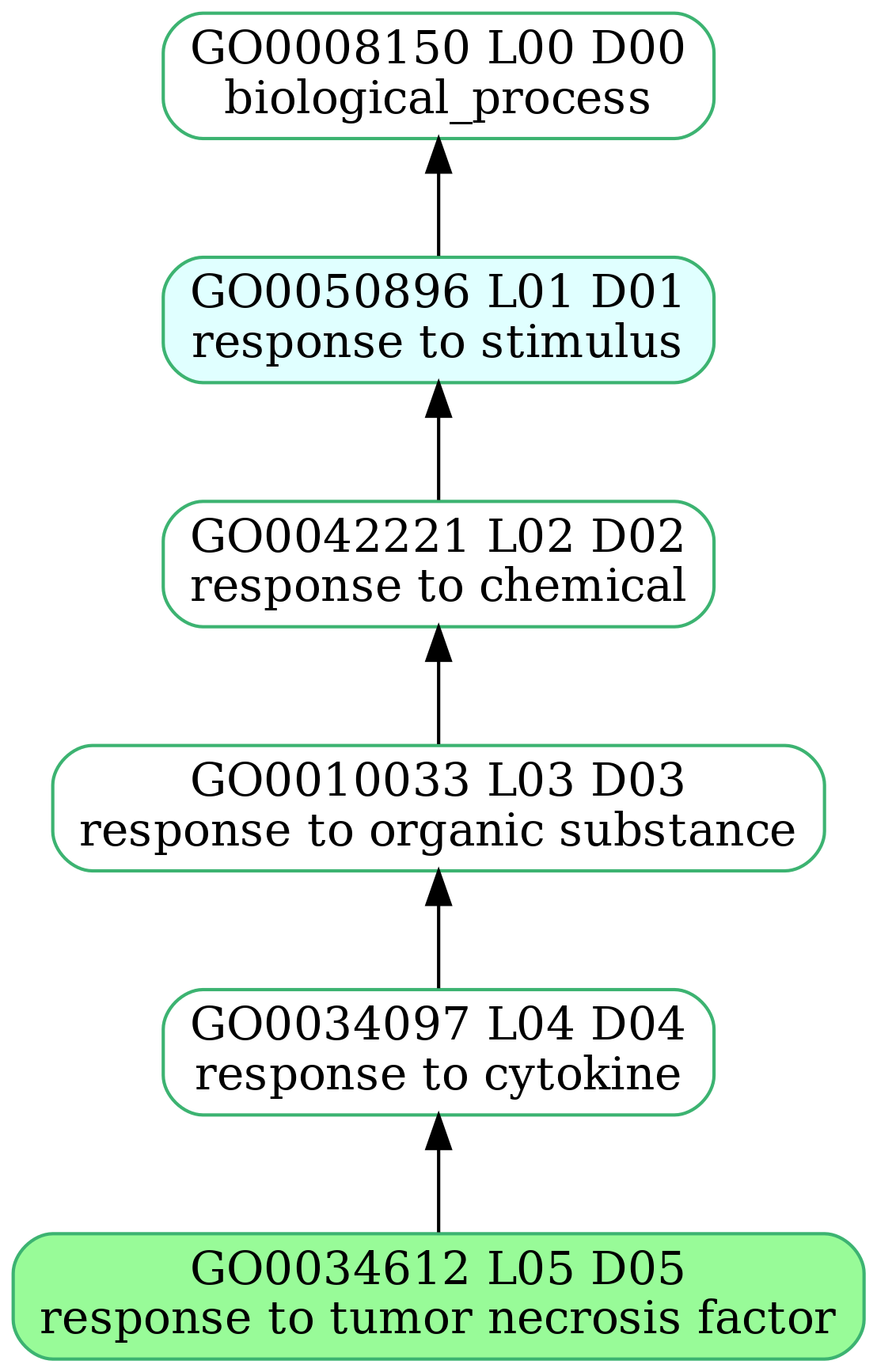
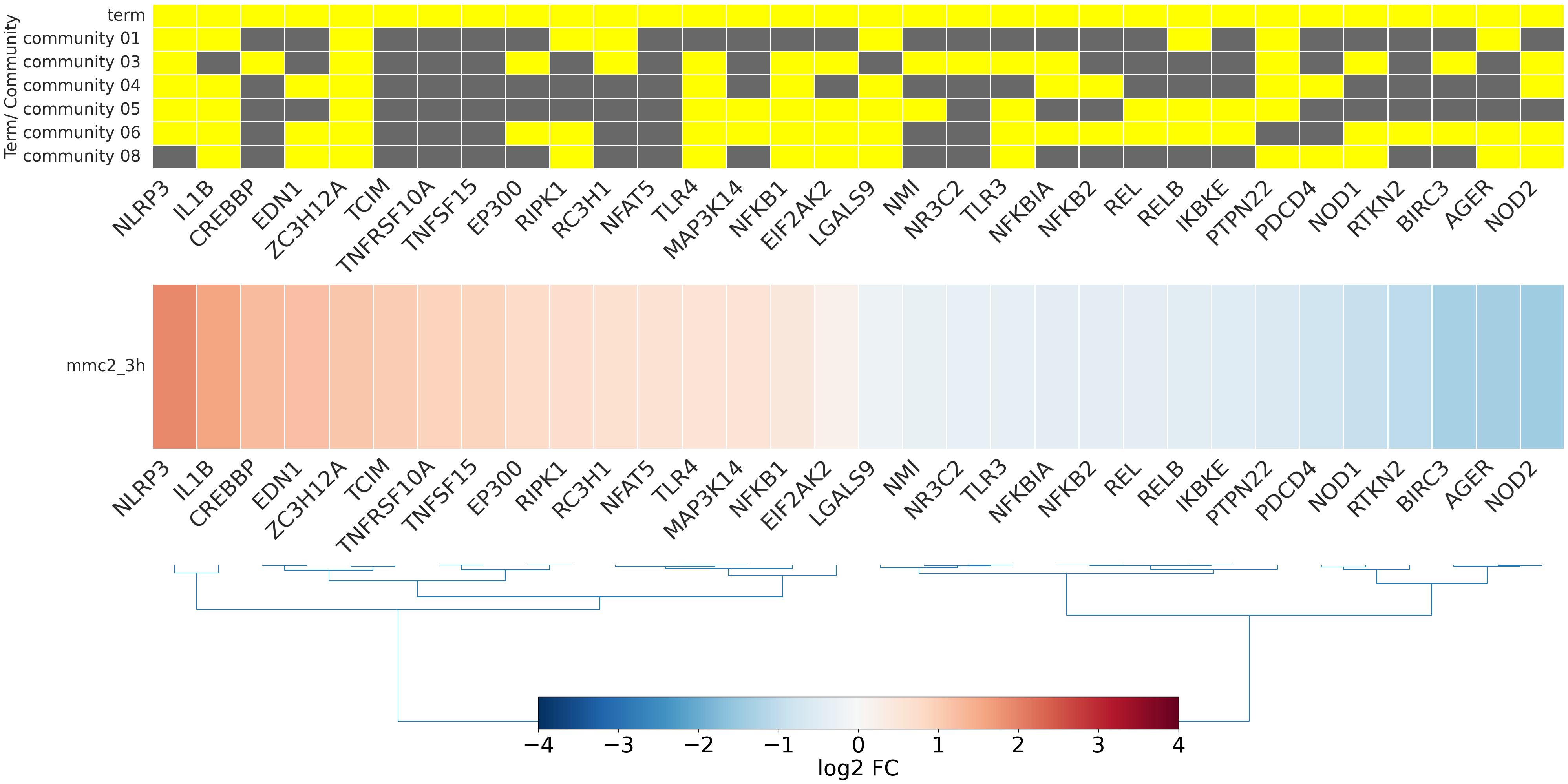
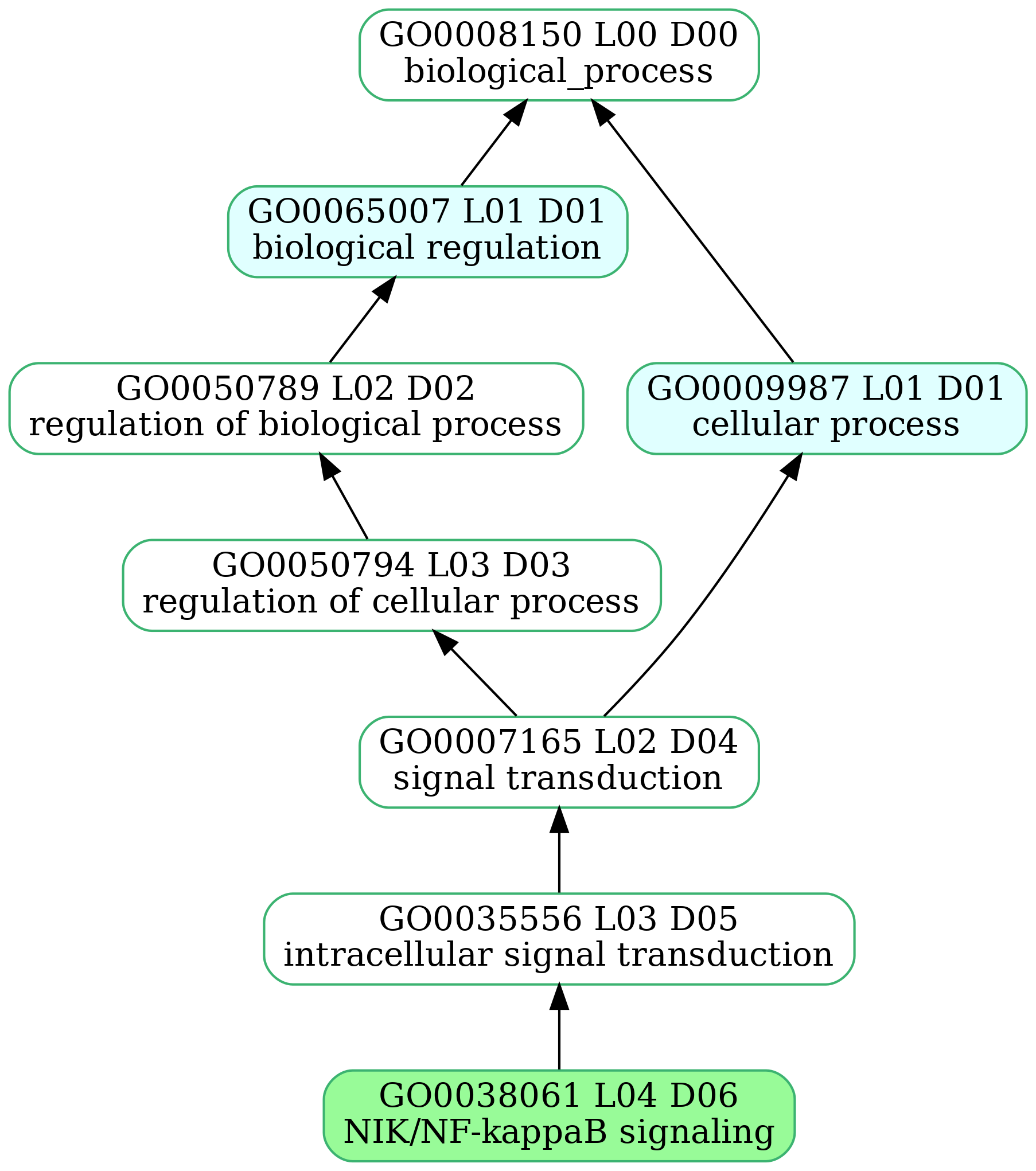
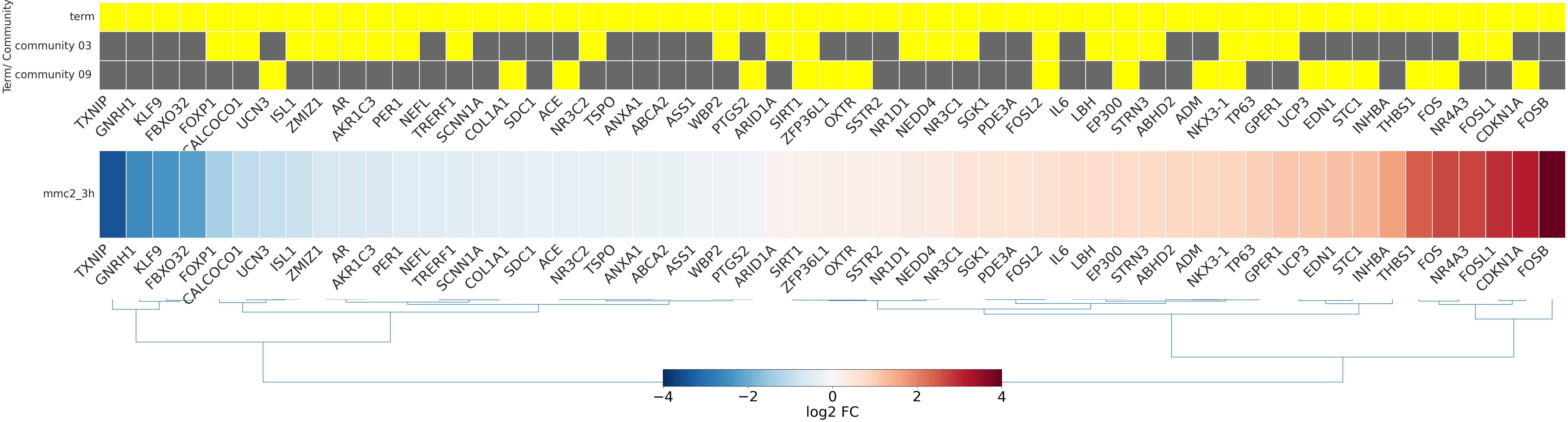
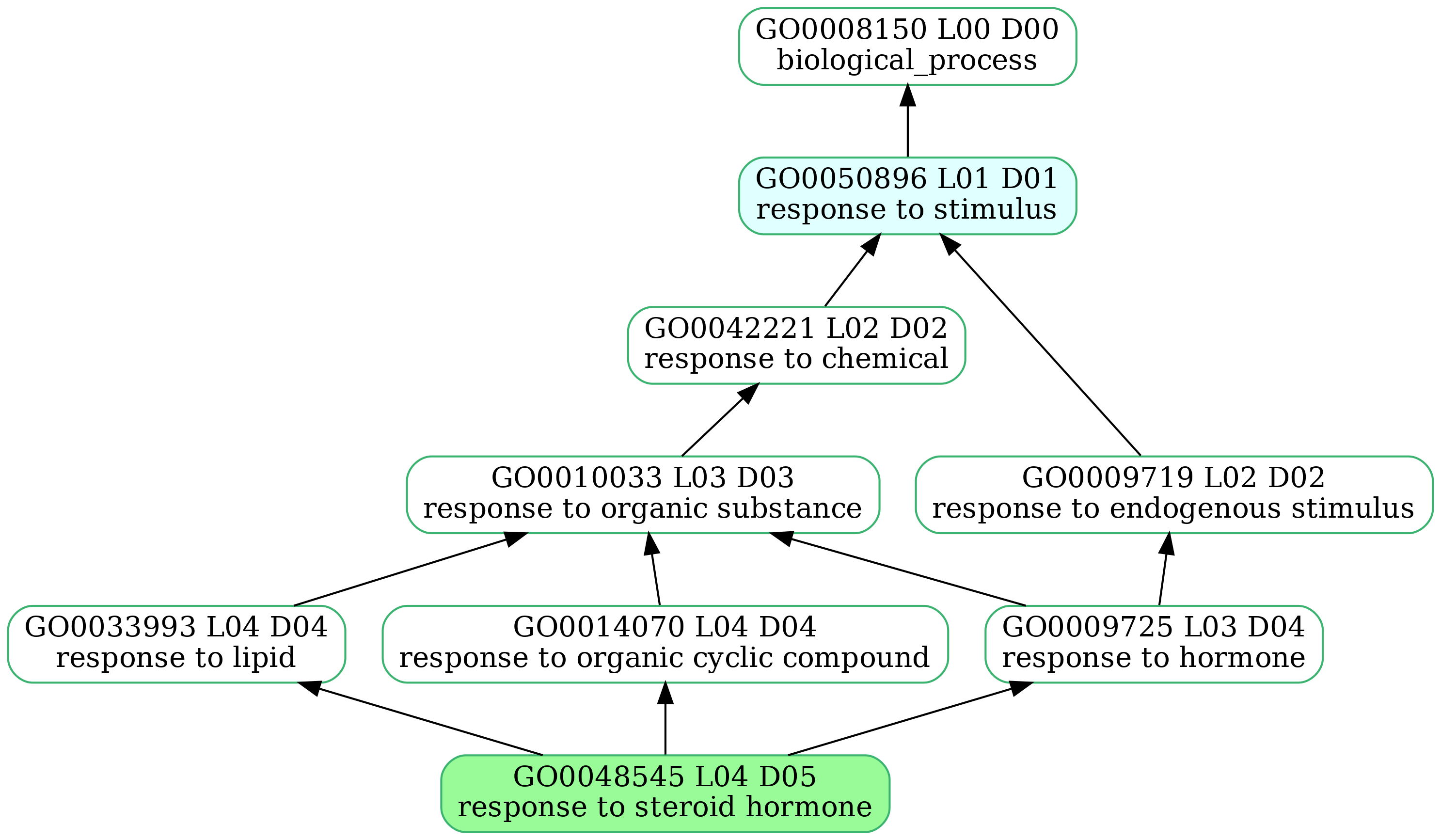
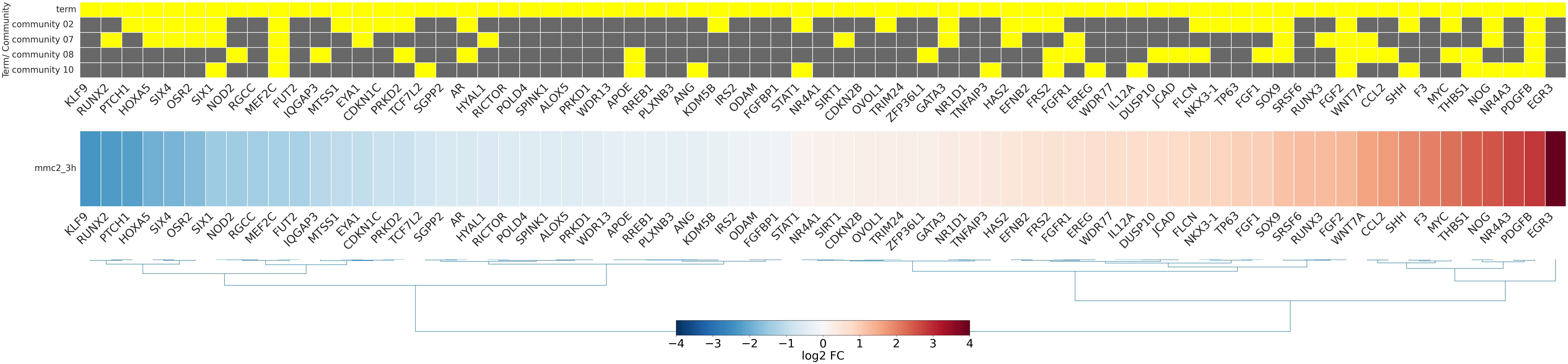
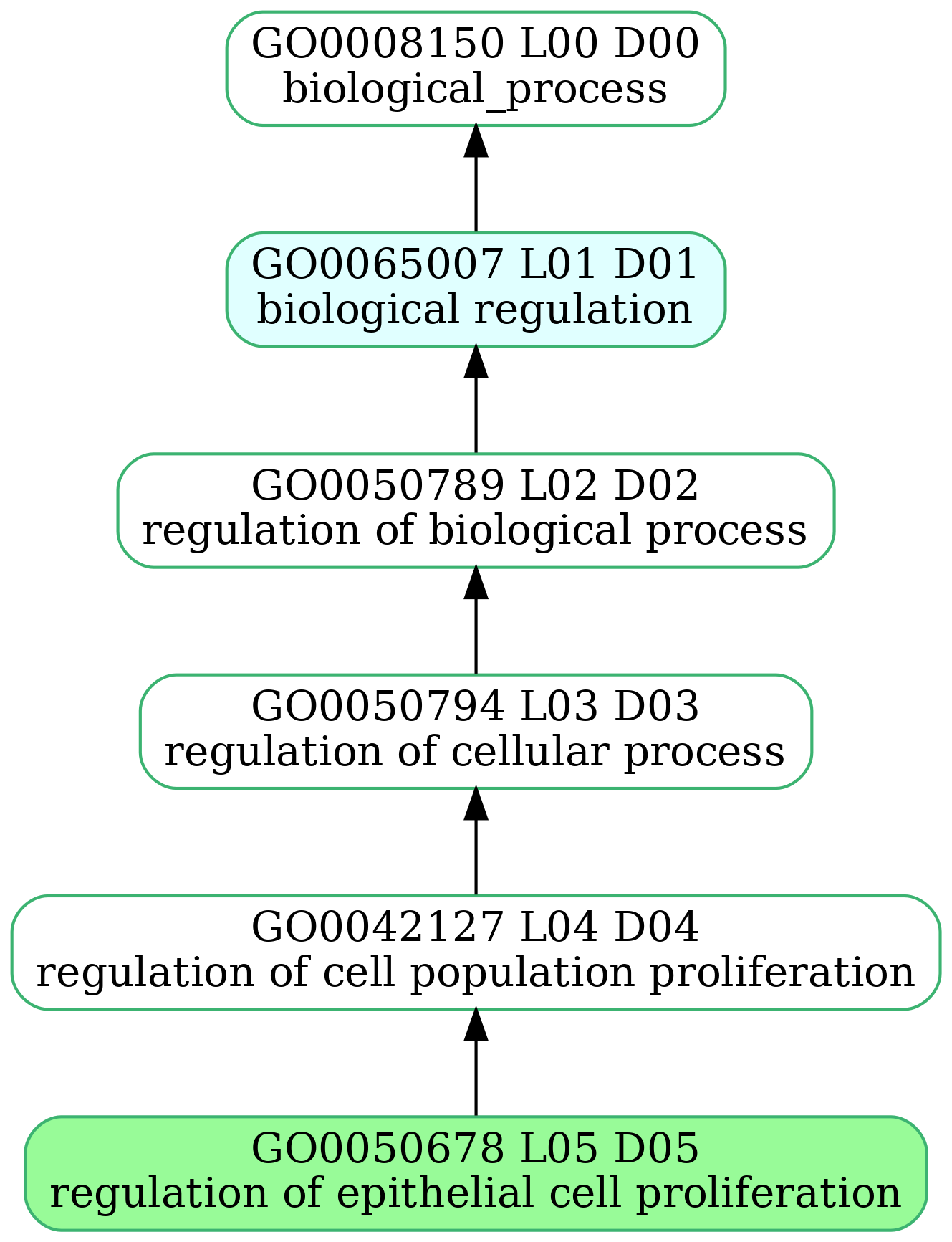
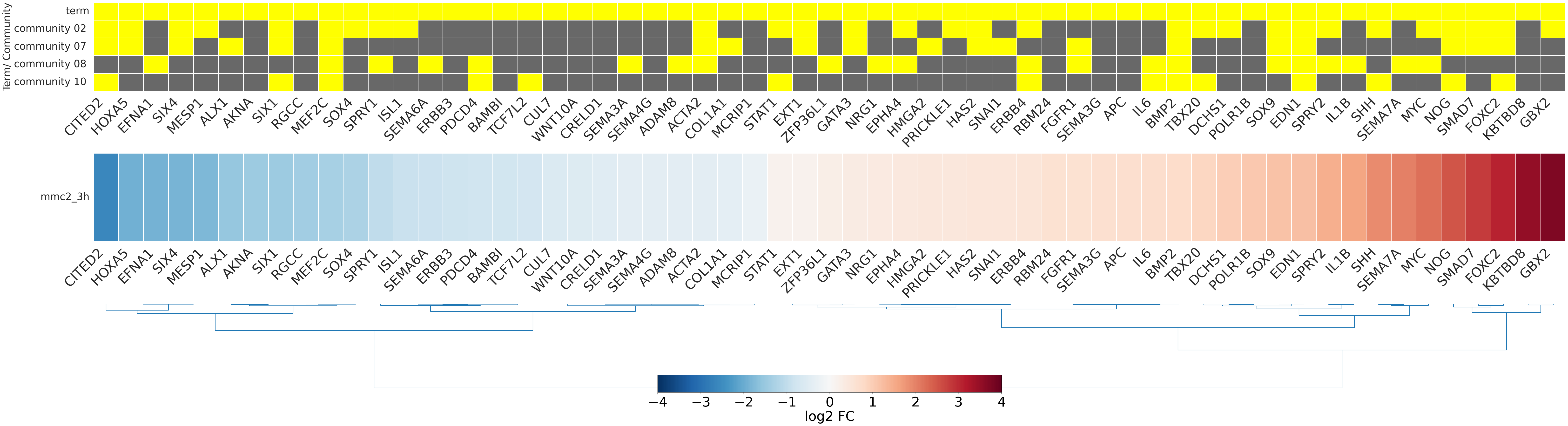
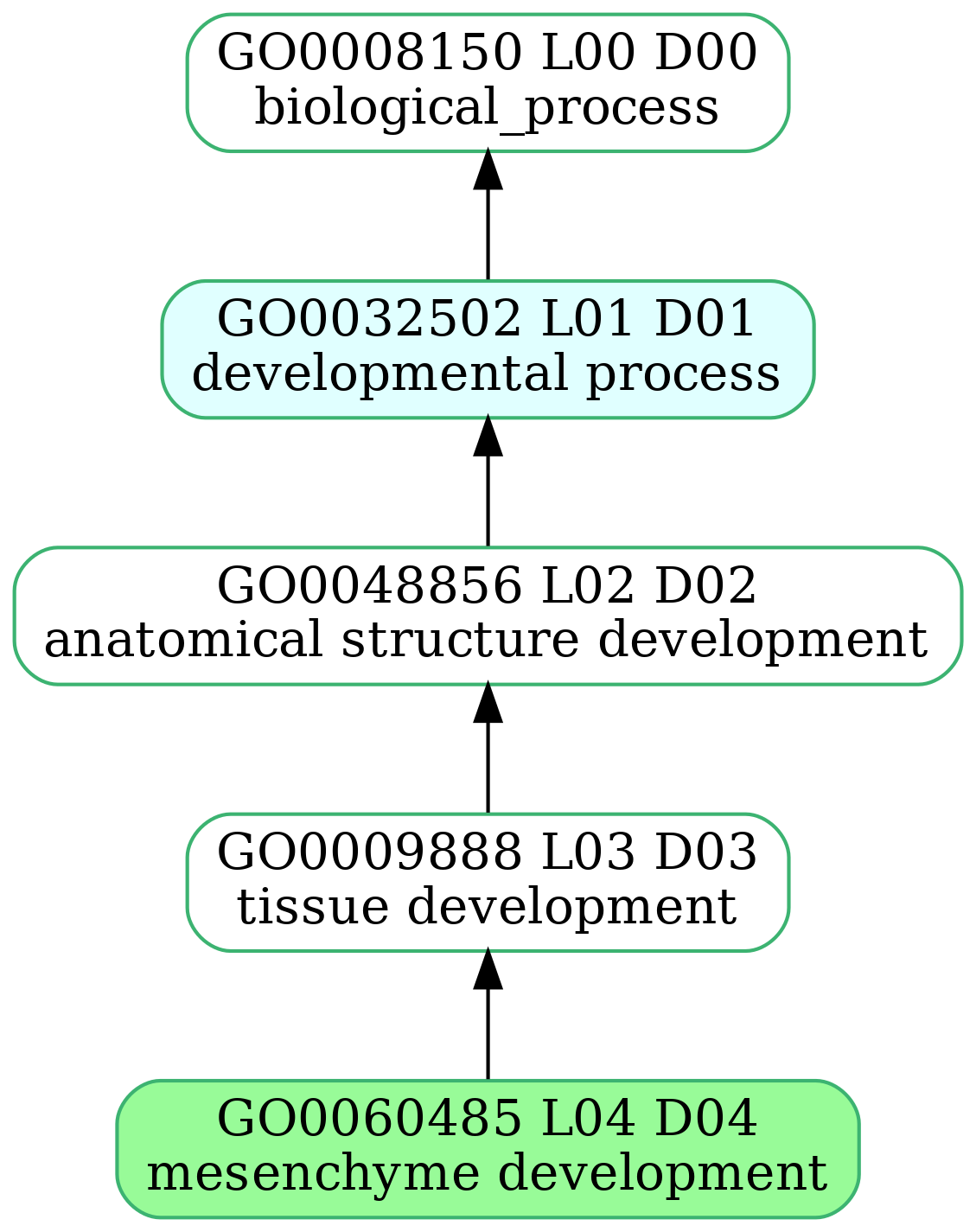
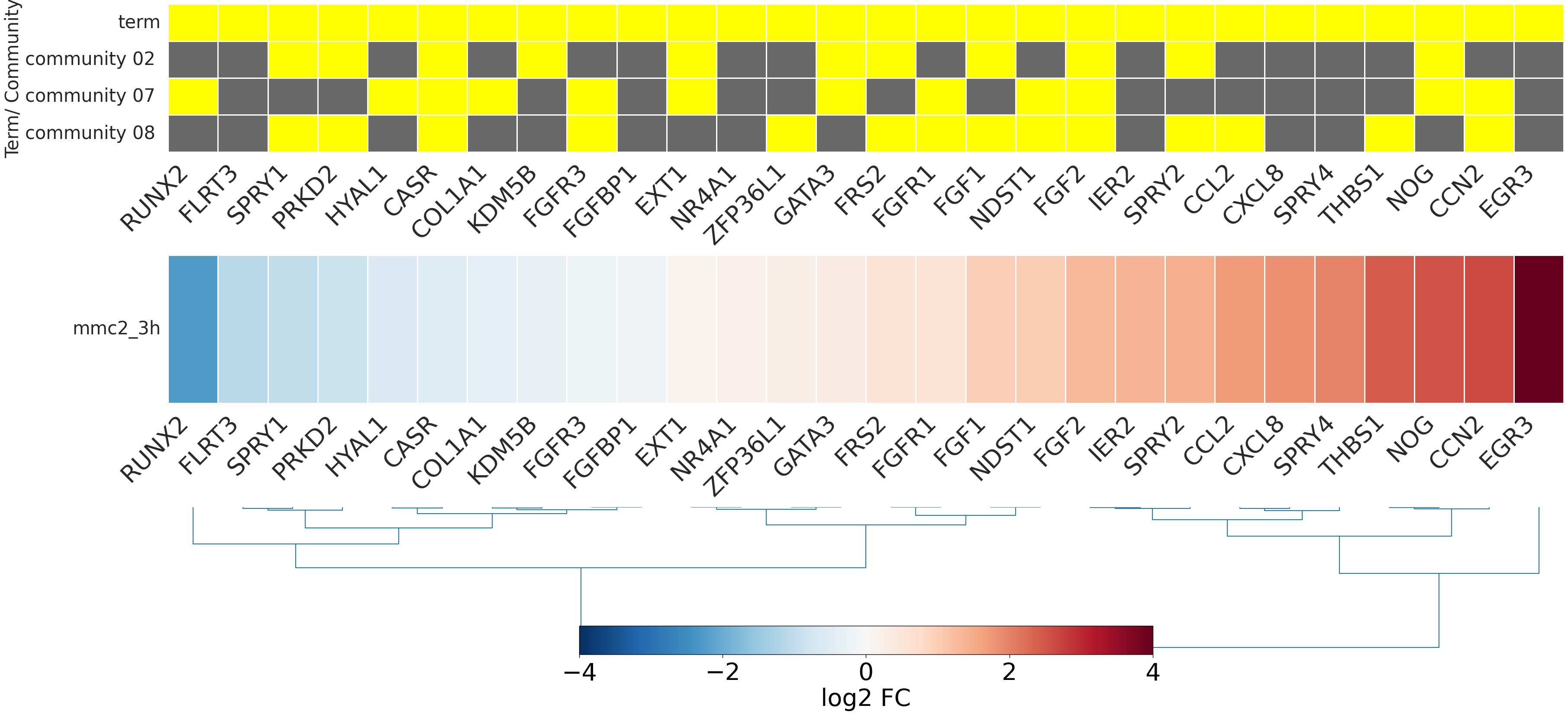
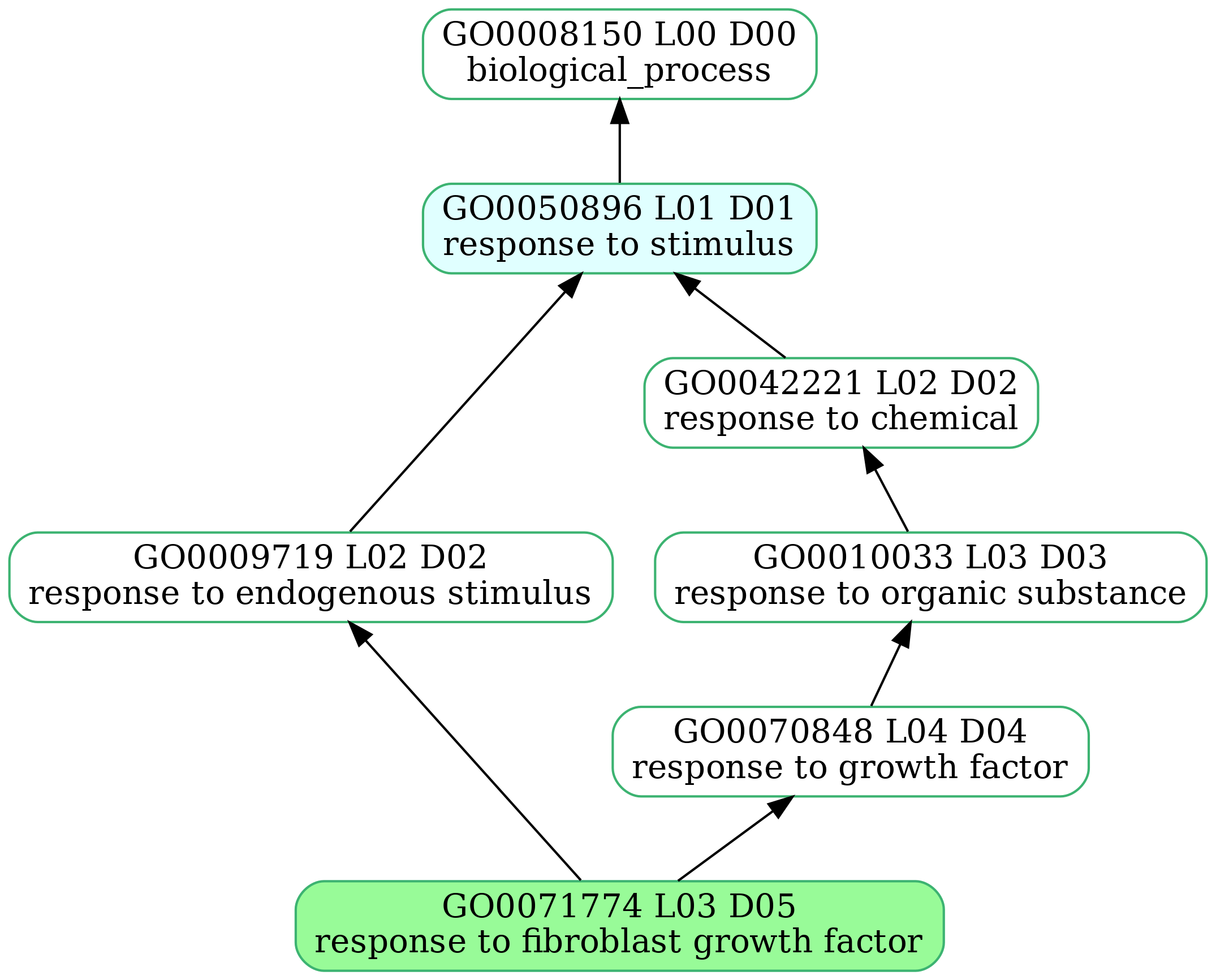
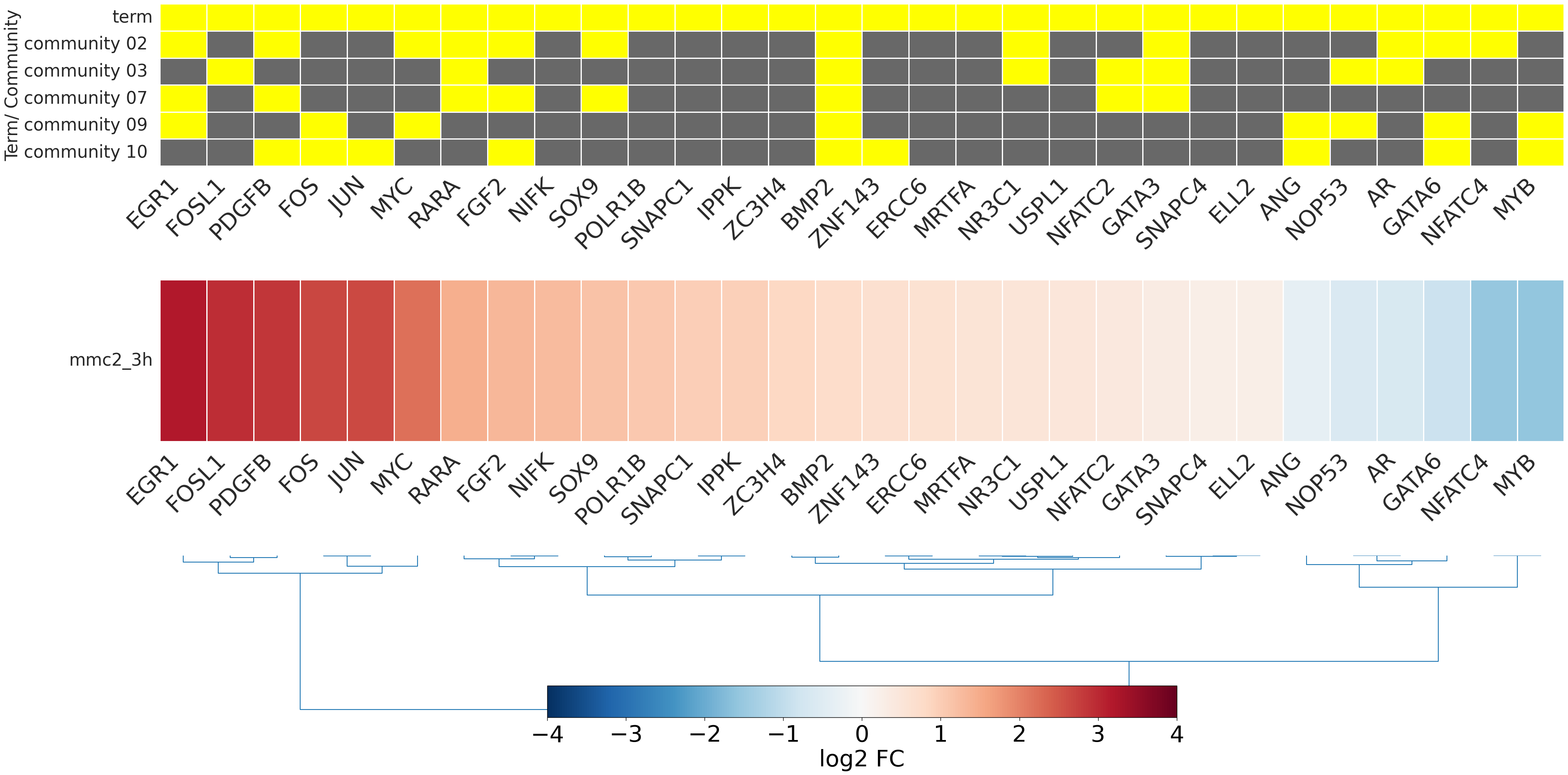
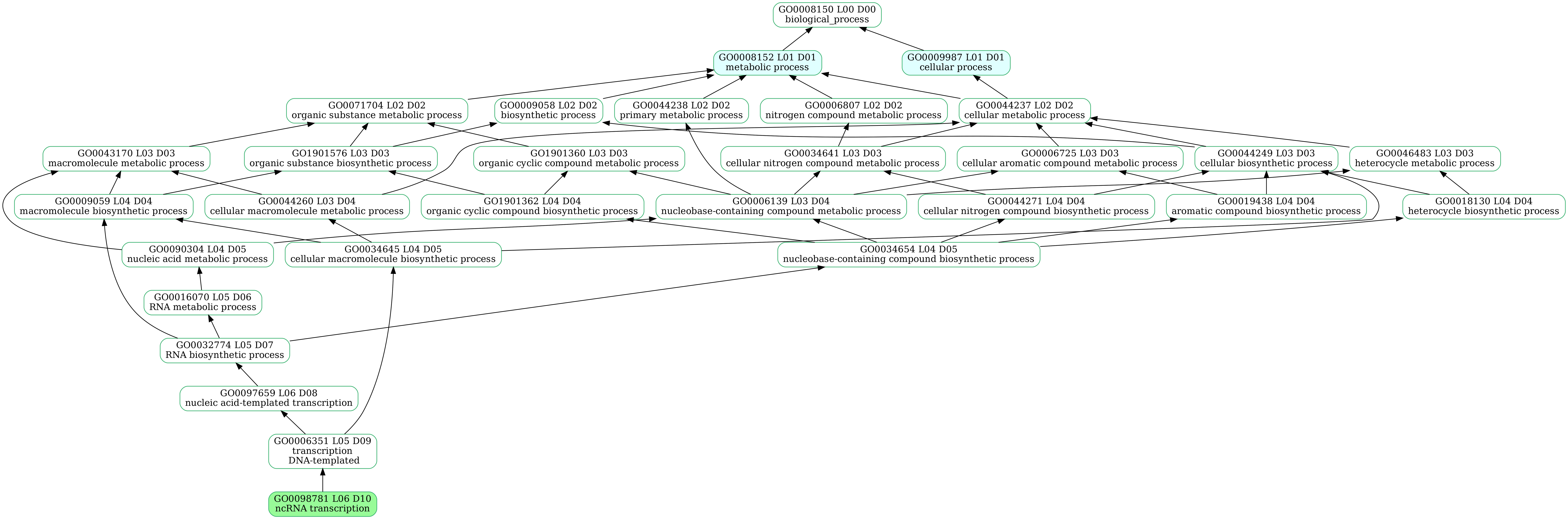
-
-
- mmc2_3h
|
Member terms:
|
|
|
|
|
|
GO:0002294 |
CD4-positive alpha-beta T cell differentiation involved in immune response
|
|
GO:0030217* |
T cell differentiation
|
|
GO:0035710 |
CD4-positive alpha-beta T cell activation
|
|
GO:0043367 |
CD4-positive alpha-beta T cell differentiation
|
|
GO:0043370 |
regulation of CD4-positive alpha-beta T cell differentiation
|
|
GO:0046631 |
alpha-beta T cell activation
|
|
GO:0046632 |
alpha-beta T cell differentiation
|
|
GO:1902107 |
positive regulation of leukocyte differentiation
|
|
GO:1903708 |
positive regulation of hemopoiesis
|
|
GO:2000514 |
regulation of CD4-positive alpha-beta T cell activation
|
|
Member terms:
|
|
|
|
|
|
GO:0002218 |
activation of innate immune response
|
|
GO:0002221 |
pattern recognition receptor signaling pathway
|
|
GO:0002753 |
cytosolic pattern recognition receptor signaling pathway
|
|
GO:0002758 |
innate immune response-activating signaling pathway
|
|
GO:0002764 |
immune response-regulating signaling pathway
|
|
GO:0030522* |
intracellular receptor signaling pathway
|
|
Member terms:
|
|
|
|
|
|
GO:0002237* |
response to molecule of bacterial origin
|
|
GO:0032496 |
response to lipopolysaccharide
|
|
GO:0071216 |
cellular response to biotic stimulus
|
|
GO:0071219 |
cellular response to molecule of bacterial origin
|
|
GO:0071222 |
cellular response to lipopolysaccharide
|
|
Member terms:
|
|
|
|
|
|
GO:0009615* |
response to virus
|
|
GO:0032479 |
regulation of type I interferon production
|
|
GO:0032606 |
type I interferon production
|
|
GO:0051607 |
defense response to virus
|
|
GO:0140546 |
defense response to symbiont
|
|
Member terms:
|
|
|
|
|
|
GO:0002720 |
positive regulation of cytokine production involved in immune response
|
|
GO:0007249 |
canonical NF-kappaB signal transduction
|
|
GO:0043122 |
regulation of canonical NF-kappaB signal transduction
|
|
GO:0051090* |
regulation of DNA-binding transcription factor activity
|
|
GO:0061081 |
positive regulation of myeloid leukocyte cytokine production involved in immune response
|
|
Member terms:
|
|
|
|
|
|
GO:0001822 |
kidney development
|
|
GO:0048645 |
animal organ formation
|
|
GO:0048754 |
branching morphogenesis of an epithelial tube
|
|
GO:0060562 |
epithelial tube morphogenesis
|
|
GO:0061138 |
morphogenesis of a branching epithelium
|
|
GO:0072001* |
renal system development
|
|
GO:0110110 |
positive regulation of animal organ morphogenesis
|
|
Member terms:
|
|
|
|
|
|
GO:0035988 |
chondrocyte proliferation
|
|
GO:0048705 |
skeletal system morphogenesis
|
|
GO:0051216 |
cartilage development
|
|
GO:0061448* |
connective tissue development
|
|
Member terms:
|
|
|
|
|
|
GO:0007254 |
JNK cascade
|
|
GO:0043410* |
positive regulation of MAPK cascade
|
|
GO:0051403 |
stress-activated MAPK cascade
|
|
GO:0070371 |
ERK1 and ERK2 cascade
|
|
Member terms:
|
|
|
|
|
|
GO:0001666 |
response to hypoxia
|
|
GO:0036293 |
response to decreased oxygen levels
|
|
GO:0070482* |
response to oxygen levels
|
|
Member terms:
|
|
|
|
|
|
GO:0033002* |
muscle cell proliferation
|
|
GO:0048660 |
regulation of smooth muscle cell proliferation
|
|
Overlapping communities:
|
|
|
|
|
|
None
|
|
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)
_circos.png)
_hierarchy.png)